Weekly Updates | Feb 11 | #2
What all was found last week? A prey that turns into a predator when temperature changes. And mtDNA cans escape into cytoplasm and trigger immune response. Whats more? Find out below:
Changing The Temperature Can Turn A Prey Into A Predator
A recent study, published in the open-access journal PLOS Biology, reveals a surprising shift in the predator-prey relationship between two bacterial species when subjected to changes in temperature. Conducted by Marie Vasse of MIVEGEC, France, and colleagues, the research highlights the impact of environmental factors on microbial interactions. While previous studies have demonstrated how ecological context, like background color or salinity, can influence predator-prey relationships, this study focuses on non-biological ecological changes, specifically temperature.
The laboratory experiments involved two bacterial species, Myxococcus xanthus and Pseudomonas fluorescens, known to engage in predator-prey dynamics. When P. fluorescens was grown at a higher temperature (32 degrees Celsius) and exposed to M. xanthus, M. xanthus acted as the predator, extensively killing P. fluorescens. However, when P. fluorescens was grown at a lower temperature (22 degrees Celsius), the roles reversed, with P. fluorescens becoming the predator, killing and obtaining nutrients from M. xanthus.
Further experiments delved into the mechanism behind this reversal, focusing on a substance released by P. fluorescens lethal to M. xanthus. The production of this substance appeared to be influenced by temperature. The study suggests that microbe-microbe killing, not typically associated with predation, may indeed result in predation, and the temperature at which bacteria grow can determine the predator-prey roles when they later meet.
The implications of this research extend to a better understanding of natural ecology and practical applications, such as optimizing the use of microbes to control others. The study emphasizes the importance of considering historical context, specifically the growth temperature of bacteria, in evaluating present predator-prey relationships. The authors highlight the significance of even small changes in ecological factors in determining microbial interactions, shedding light on a previously underappreciated aspect of microbe-microbe killing and predation.
References:
Chill To Kill: How Cooling Turns Prey Bacteria Into Predators (scitechdaily.com)
Killer prey: Ecology reverses bacterial predation” by Marie Vasse, Francesca Fiegna, Ben Kriesel and Gregory J. Velicer, 23 January 2024, PLOS Biology
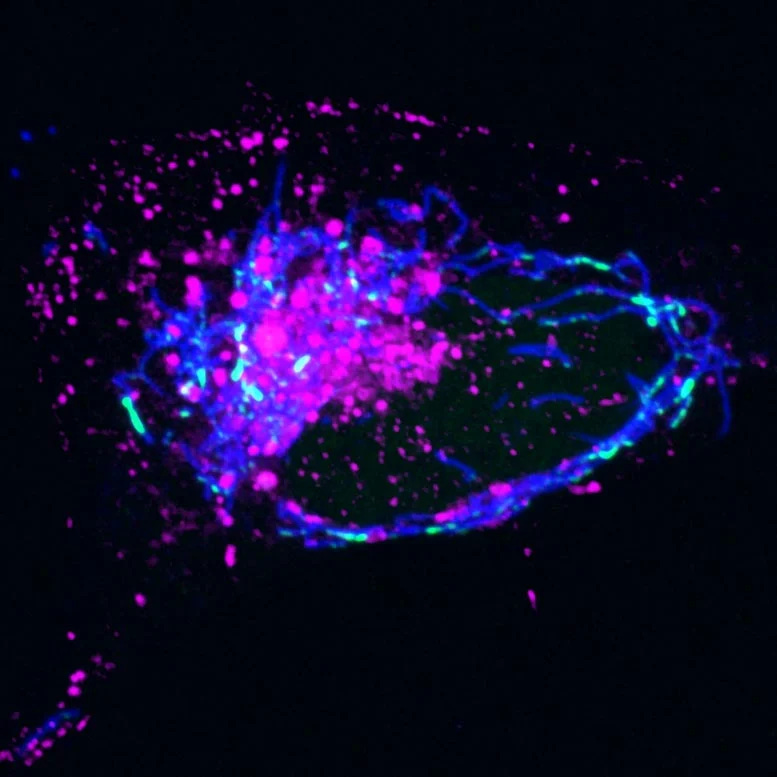
When mtDNA Leaks Into Cytoplasm
Researchers from the Salk Institute and UC San Diego have uncovered a novel mechanism for removing malfunctioning mitochondrial DNA (mtDNA) from inside to outside the mitochondria, triggering an immune response that leads to inflammation. The study, published in Nature Cell Biology, reveals that when mtDNA escapes mitochondria, it can activate a cellular pathway that promotes inflammation to eliminate the foreign DNA. The findings offer potential therapeutic targets to disrupt this inflammatory pathway, providing new avenues for reducing inflammation in conditions such as aging and diseases like lupus or rheumatoid arthritis.
Fig: Magenta - Endosomes; Blue - Mitochondria | Image Credit : Salk Institute
Senior and co-corresponding author Professor Gerald Shadel explains that the study traced the steps of the pathway for moving mtDNA out of mitochondria using imaging and cell biology approaches. The researchers discovered a malfunction in mtDNA replication, causing nucleoids to accumulate inside mitochondria. The cell responds by transporting these nucleoids to endosomes, which eventually leak, releasing mtDNA into the cell. The cell recognizes the foreign mtDNA, triggering the cGAS-STING pathway and promoting inflammation.
Laura Newman, the first author of the study, notes that the breakthrough came when they realized the mtDNA was inside a membrane structure, identified as an endosome, after leaving mitochondria. The researchers hope to explore further details of this mtDNA-disposal and immune-activation pathway, including the biological circumstances that initiate it and its downstream effects on human health. Therapeutically targeting this pathway could offer a novel approach to reduce inflammation during aging and various diseases.
Reference:
Multicellularity In Plants Might Have Evolved Earlier Than We Thought
A team of scientists led by the University of Göttingen has delved into the evolutionary history of complex green organisms, particularly land plants and various green algae. Their focus centred on Klebsormidiophyceae, a class of green algae renowned for its adaptability to diverse habitats across the globe. In their research, the scientists conducted extensive sampling in various environments, such as streams, rivers, bogs, urban walls, and more, aiming to create a comprehensive global distribution map for Klebsormidiophyceae. This approach underscored the adaptability, ecological significance, and hidden diversity of these algae.
One of the challenges the researchers faced was resolving phylogenetic relationships using traditional markers. To overcome this, they employed phylogenomics, a powerful method that involves reconstructing evolutionary history by considering whole genomes or large genome fractions. This innovative approach allowed them to create a new phylogenomic tree of life for Klebsormidiophyceae, dividing it into three orders. The results of their study provided valuable insights into the ancient ancestry of Klebsormidiophyceae, revealing a multicellular ancestor that thrived millions of years ago and began splitting into distinct branches over 800 million years ago.
References:
New Research Reveals That Complex Green Organisms Emerged a Billion Years Ago (scitechdaily.com)
Phylogenomic insights into the first multicellular streptophyte” by Maaike J. Bierenbroodspot, Tatyana Darienko, Sophie de Vries, Janine M.R. Fürst-Jansen, Henrik Buschmann, Thomas Pröschold, Iker Irisarri and Jan de Vries, 19 January 2024, Current Biology
How Different Amino Acids Affect Cell Growth By Activating TORC1?
Scientists at Osaka University have made a significant breakthrough in understanding the mechanism by which amino acids, particularly cysteine, activate TORC1, a master regulator protein crucial for cell growth in yeast. Amino acids are essential components for life, sourced from our diet, and utilized by the body to create proteins vital for various processes, including growth and development. When amino acids are present, TORC1 is activated, leading to protein synthesis and cell growth. The study, published in Cell Reports, reveals that cysteine is detected by a protein called Pib2, which then binds to TORC1, activating it and stimulating the synthesis of proteins and lipids, thereby promoting cell proliferation.
The research also discovered that cysteine is not the only amino acid influencing TORC1. All 20 amino acids were found to have varying effects on TORC1 through two pathways: Pib2 and Gtr. The study aimed to understand how each amino acid utilizes these pathways to impact TORC1 activation. The findings indicate that some amino acids primarily use the Pib2 pathway, others use the Gtr pathway, and some can use either or both pathways. This deepens the understanding of how amino acids control cell growth and autophagy, and how their detection plays a crucial role in these processes.
The implications of this research extend to human health, as faulty TORC1 function in humans has been linked to diseases such as cancer, diabetes, and dementia. Understanding how TORC1 is switched on and off, along with the detection of each amino acid, could pave the way for new treatments for these diseases. The study provides valuable insights that may contribute to advancements in medical research and the development of potential therapeutic interventions for TORC1-related disorders.
References:
Scientists Elucidate Key Mechanism to Cell Growth (scitechdaily.com)
Pib2 is a cysteine sensor involved in TORC1 activation in Saccharomyces cerevisiae” by Qingzhong Zeng, Yasuhiro Araki and Takeshi Noda, 20 December 2023, Cell Reports
Trophoblast Stem (TS) Cell-Based Organoid Model of the Human Placental Barrier
In a recent article published in Nature Communications, researchers from Tokyo Medical and Dental University (TMDU) have made significant strides in developing a trophoblast stem (TS) cell-based organoid model of the human placental barrier. The human placenta, crucial during pregnancy for various roles such as hormone production and nutrient processing, also acts as a barrier to shield the developing fetus from external harmful substances. However, certain drugs can breach this protective barrier, necessitating a reliable in vitro model for research.
The TMDU team faced challenges in replicating the structural nature of placental villi, which are essential for the barrier's function. Previous methods, including cell lines and other approaches, proved inadequate, and primary placental cells were difficult to maintain in culture. The researchers turned to TS cells, which have the potential to differentiate into various placental cells, to develop an effective in vitro model.
The team generated trophoblast organoids, three-dimensional cell models that better mimic the structural and biological details of organs. By optimizing culture conditions and creating a model with a single layer of cells called syncytiotrophoblasts, the researchers successfully displayed the barrier function they aimed to replicate. This organoid model, with its ability to assess the translocation of compounds through the barrier layer, addresses previous difficulties in laboratory-based assessments of placental physiology.
Dr. Takeshi Hori, the lead author of the study, emphasizes the model's potential for understanding general placental biology and drug toxicity. The researchers designed the model for easy cell culture and microscopic evaluation, making it a valuable tool for studying placental development and evaluating compound transfer rates and toxicity levels. This advancement is particularly critical in drug development, helping to avoid damage to the placenta and potential harm to the fetus. The TS cell-based organoid model holds promise for advancing research in placental biology and contributing to safer drug development processes.
Reference: